Smart Seq
Echo SystemEnhanced SMARTSeq v4 for RNA Sequencing Introduction As the cost of sequencing has continued to decline by orders of magnitude in the past ten years, scientists are enabled to ask deeper and more complex questions about transcriptomics Consequently, the variety, quantity, and demands of RNA sequencing experiments have all increased.

Smart seq. Note these data were included in the subsequent “Human Multiple Cortical Areas SMARTseq” data set, which can be found here This data set includes singlenucleus transcriptomes from 16,281 total nuclei collected from human primary visual cortex and anterior cingulate cortex (ACC;. We developed Perturbseq to combine a pooled CRISPR screen with scRNAseq by encoding the identity of the perturbation on an expressed guide barcode (GBC) (Figures 1 and S1)We first infect cells with a pool of lentiviral constructs that encode singleguide RNAs (sgRNAs) (Figure 1A)Here, and in a companion study (Adamson et al, 16 this issue of Cell), we designed and used a CRISPR. Overview of SmartSeq protocol SmartSeq cDNA illumina Nextera XT • Fragment cDNA and add sequencing adapters • Pairedend sequencing recommended (unstranded library) • ~3040 million reads per sample recommended.
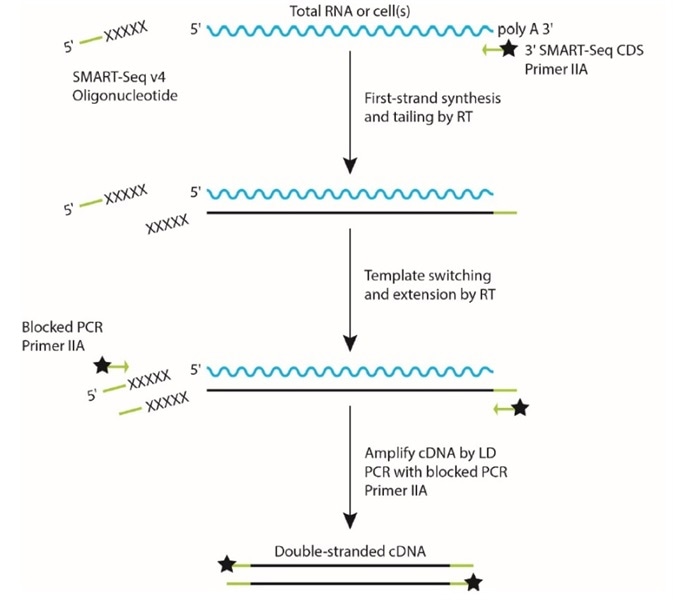
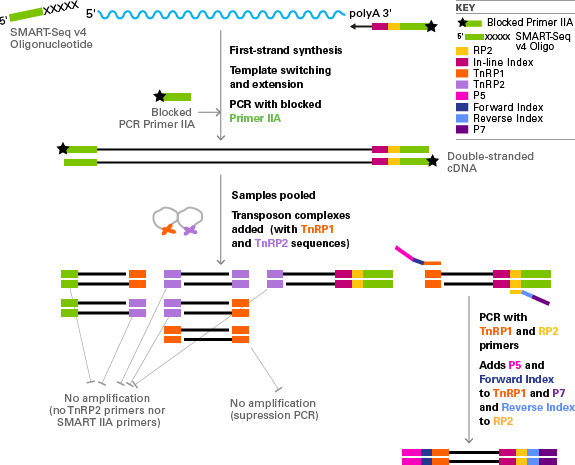
SMARTSeq v2, a method developed by Simone Picelli et al at the Karolinska Institute, has become a ubiquitous method for singlecell and population RNA sequencing Here, we demonstrate miniaturization of this process on human brain total RNA utilizing the Echo 525 Liquid Handler We show that this enhanced process can effectively reduce. SMARTSeq v4 Reagent Kit for the SMARTer Apollo System (Cat No ) is designed to generate highquality, fulllength cDNA directly from 1–1,000 cells or 10 pg–10 ng of total RNA. All samples were processed with the Smartseq library construction method for fulllength coverage of individual transcripts (Picelli et al, 13) This resulted in a compendium of seven embryonic lineages from six developmental stages, spanning zygote, fourcell, eightcell, compacted morula, and early and late ICM.
Single Molecule, RealTime (SMRT) Sequencing is the core technology powering our longread sequencing platforms This innovative approach was the first of its kind and is now a proven technology used in all fields of life science PacBio Sequencing – How it Works. The Smartseq2 method exploits the socalled SMART reaction (Switching Mechanism At the end of the 5′end of the RNA Transcript), developed by scientists at CLONTECH several years ago (Chenchik et al, 1996;. Please do not hesitate to contact us filling the form below.
The protocol below uses the SMARTer Stranded Total RNASeq Kit, however another option for cDNA generation when starting with lowinput is the Clontech ‘SMART Seq HT’ for Highthroughput singlecell mRNAseq This is an excellent kit for preparing cDNA very low amounts of RNA (as little as 1100 cells). Archives smartseq FORMULATRIX® Present Validated SMARTSeq Protocol for Miniaturized cDNA Synthesis Reactions October 11, 17 Leave a comment 4,026 Views. PacBio Single Molecule, RealTime (SMRT) Sequencing powers our longread sequencing platforms SMRT Sequencing offers long reads, high accuarcy, uniform coverage, singlemolecule resolution, and epigenetics to drive discovery in life science.
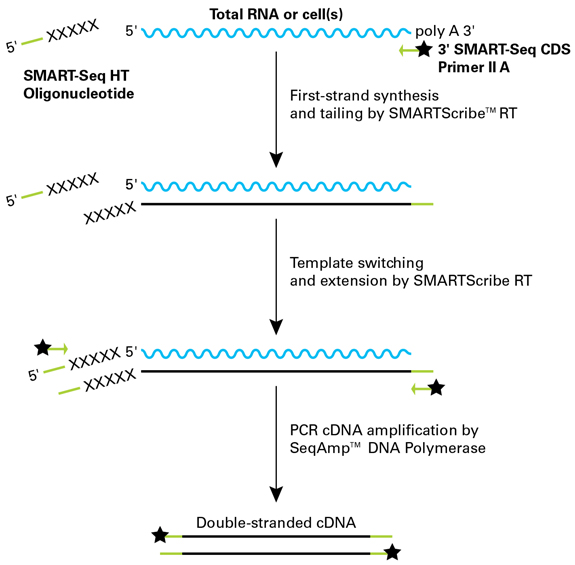
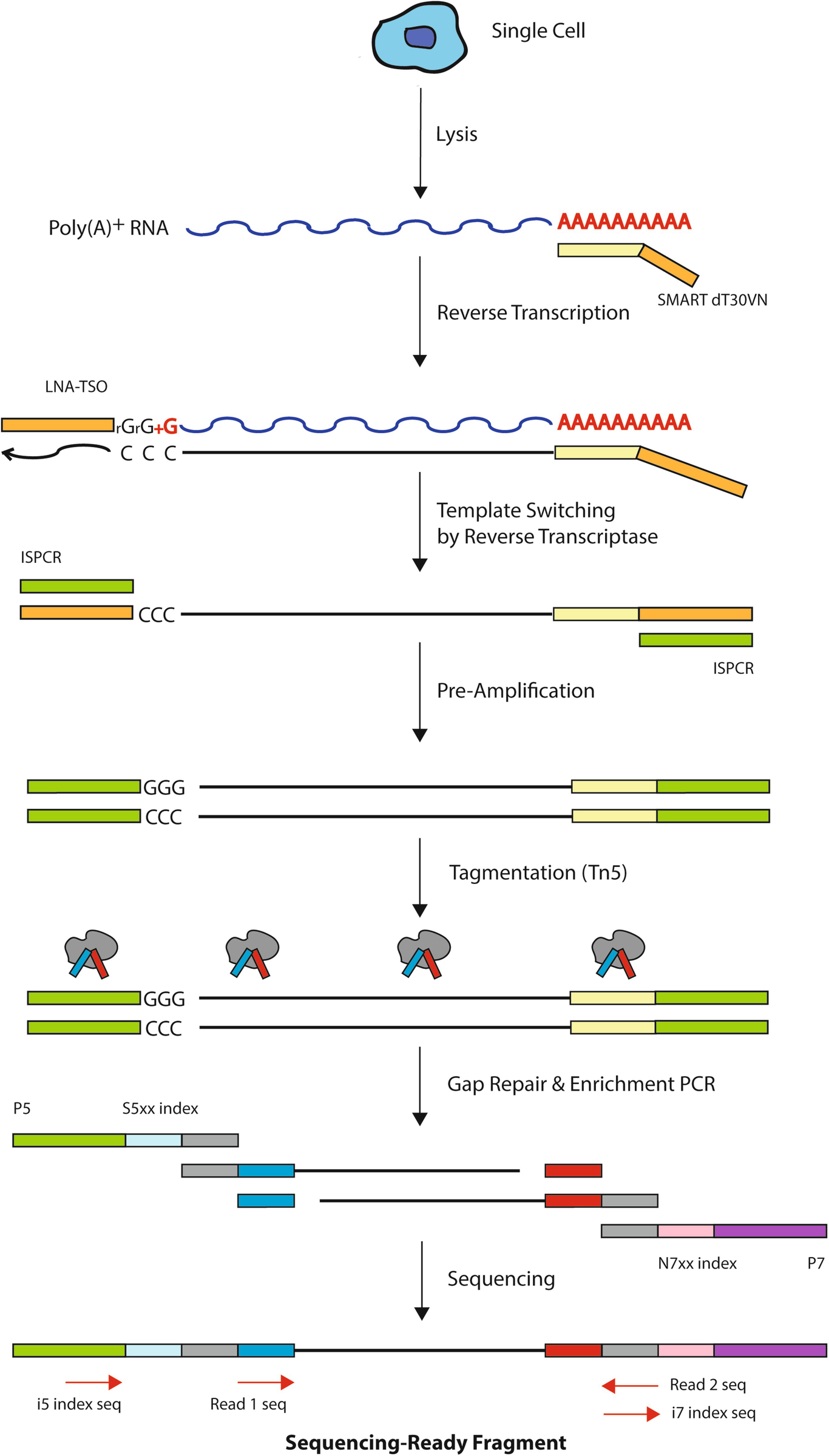
SMARTSeq HT Kit Streamlined singlecell mRNAseq with the SMARTSeq HT Kit The SMARTSeq HT Kit (SSHT) is an automationfriendly kit that uses oligo(dT) priming to generate highquality, fulllength cDNA directly from 1–100 cells or 10 pg–1 ng of total RNA. SmartSeq Cells are lysed, and the RNA is hybridized to an oligo (dT)containing primer The first strand of the cDNA is synthesized with the addition of a few untemplated C nucleotides This poly (C) overhang is added exclusively to fulllength transcripts. Note these data were included in the subsequent “Human Multiple Cortical Areas SMARTseq” data set, which can be found here This data set includes singlenucleus transcriptomes from 15,928 total nuclei derived from both frozen and neurosurgical human brain specimens, to survey cell type diversity in the human middle temporal gyrus ()Nuclei from 8 human tissue donors ranging in age.
For the SMARTSeq Stranded Kit, the Ultra Low Input workflow described in the user manual was followed by pooling of eight samples according to Appendix A of the user manual For the SMARTer Stranded Total RNASeq Kit v2 – Pico Input Mammalian, the cells were also processed as described for the SMARTSeq Stranded Kit, but using the reagents. Figure 1 MANTIS liquid handler by Formulatrix Image Credit FORMULATRIX Smartseq3 is a unique quantitative fulllength scRNAseq protocol Smartseq3 uses the established 96 & 384 well. Bioinformatics services We offer a complete service of custom bioinformatics analysis, database customization and implementation, wet and dry support for RNASeq, metagenomics and SNPs identification.
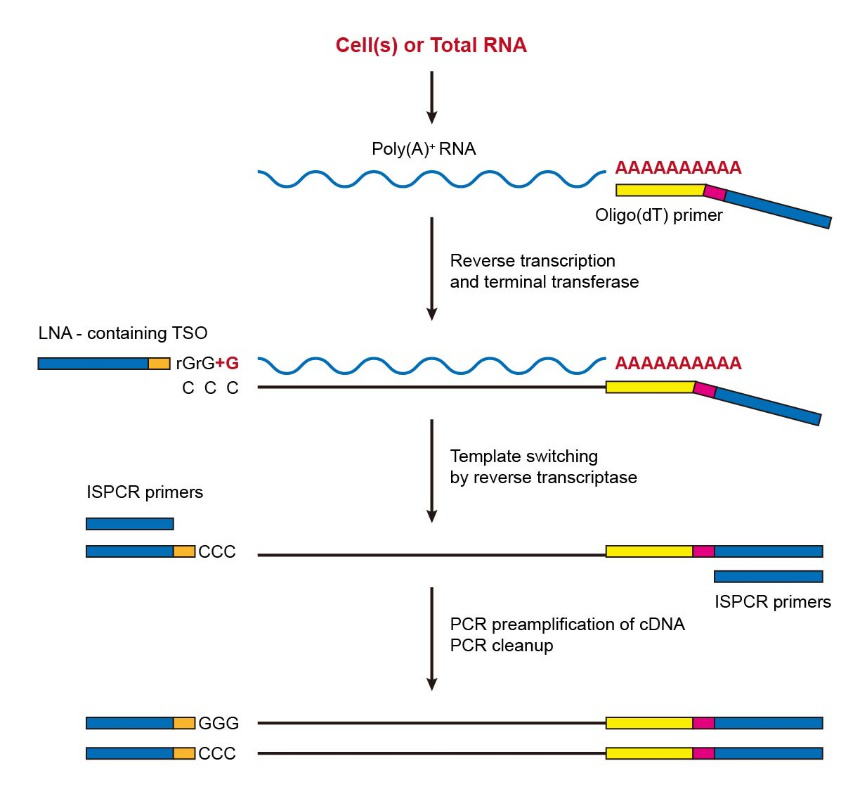
Smartseq3 Protocol method by Michael HagemannJensen. Dr Nathan Archer presents his work regarding optimization of SMARTSeq cDNA synthesis reactions as part of a webinar hosted by TakaraBio and Formulatrix Cl. SMARTseq / SMARTseq2 / SMARTseq3 The protocols of SMARTseq and SMARTseq2 are almost the same SMARTseq2 is an improved version of SMARTseq The authors performed 457 optimisation experiments to test conditions Two key parameters are (1) exchanging the last guanylate for a locked nucleic acid (LNA) at the 3' end of TSO.
SMARTseq2 is an improved version of SMARTseq The authors performed 457 optimisation experiments to test conditions Two key parameters are (1) exchanging the last guanylate for a locked nucleic acid (LNA) at the 3' end of TSO. Largescale sequencing of RNAs from individual cells can reveal patterns of gene, isoform and allelic expression across cell types and states11 However, current singlecell RNAsequencing (scRNAseq) methods have limited ability to count RNAs at allele and isoform resolution, and longread sequencing techniques lack the depth required for largescale applications across cells22,33. Recently, highthroughput sequencing combining bisulfite treatment (BisulfiteSeq) have been used to generate DNA methylomes from a wide range of human tissue/cell types at a genomewide perspective.
SMARTSeq v2, a method developed by Simone Picelli et al at the Karolinska Institute, has become a ubiquitous method for singlecell and population RNA sequencing Here, we demonstrate miniaturization of this process on human brain total RNA utilizing the Echo 525 Liquid Handler We show that this enhanced process can effectively reduce. SmartSeq was developed as a singlecell sequencing protocol with improved read coverage across transcripts (Ramskold et al, 12) Complete coverage across the genome allows the detection of alternative transcript isoforms and SNPs There are 2 versions of SmartSeq SmartSeq and Smartseq2. Here, we describe Smartseqtotal, a method capable of assaying a broad spectrum of coding and noncoding RNA from a single cell Built upon the templateswitch mechanism, Smartseqtotal bears the key feature of its predecessor, Smartseq2, namely, the ability to capture fulllength transcripts with high yield and quality.
Zhu et al, 01). About Seurat Seurat is an R package designed for QC, analysis, and exploration of singlecell RNAseq data Seurat aims to enable users to identify and interpret sources of heterogeneity from singlecell transcriptomic measurements, and to integrate diverse types of singlecell data. About Seurat Seurat is an R package designed for QC, analysis, and exploration of singlecell RNAseq data Seurat aims to enable users to identify and interpret sources of heterogeneity from singlecell transcriptomic measurements, and to integrate diverse types of singlecell data.
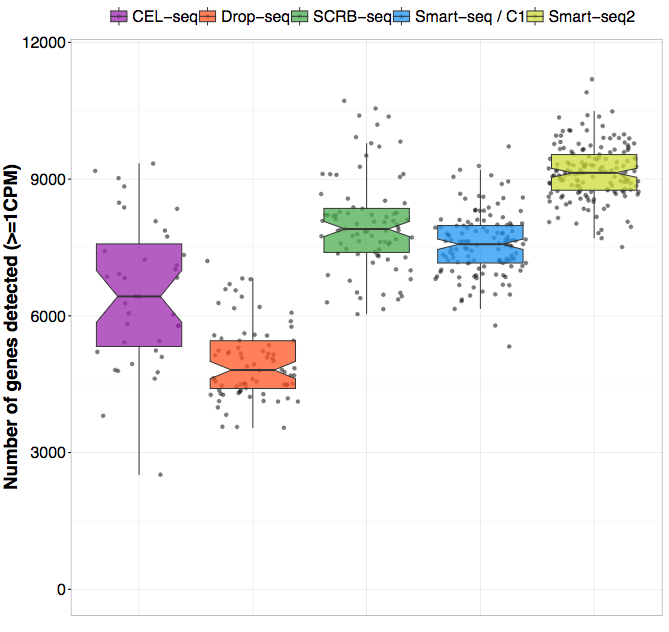
SMARTSeq HT Kit Streamlined singlecell mRNAseq with the SMARTSeq HT Kit The SMARTSeq HT Kit (SSHT) is an automationfriendly kit that uses oligo(dT) priming to generate highquality, fulllength cDNA directly from 1–100 cells or 10 pg–1 ng of total RNA. A previous study demonstrated that Smartseq2 can detect a bigger number of expressed genes than other scRNAseq technologies including CELseq2 (Hashimshony et al, 16), MARSseq (Jaitin et al, 14), Smartseq (Ramskold et al, 12), and Dropseq protocols (Ziegenhain et al, 17). Optimization & Evaluation of SMARTSeq® v4 kit for Low Input RNA Sequencing An improved strategy for expression profiling via RNA sequencing from limiting amounts of RNA Inferring the gene expression status of patients’ tumors via analysis of the residual tumor material is often challenged by the small quantity of tumor cells or tissues.
The SMARTSeq v4 Kit (SSv4) and the SMARTSeq v4 PLUS Kit (SSv4 PLUS) are the fourth generation of our SMARTer ultralow solutions and our most sensitive kits for ultralow inputs These kits use oligo (dT) priming to generate highquality, fulllength cDNA directly from multiple intact cells (up to 1,000 cells) or 10 pg–10 ng of total RNA. For the past several decades, due to technical limitations, the field of transcriptomics has focused on population‐level measurements that can mask significant differences between. Allen Cell Types Database Overview Cell Feature Search RNASeq Data Documentation Acknowledgements Help Cell Feature Search RNASeq Data Documentation Acknowledgements Help.
Different highthroughput methods for singlecell RNAseq have been introduced that vary in coverage, sensitivity and multiplexing ability We recently introduced Smartseq for transcriptome analysis from single cells, and we subsequently optimized the method for improved sensitivity, accuracy and fulllength coverage across transcripts. Let's Get In Touch!. PacBio Single Molecule, RealTime (SMRT) Sequencing powers our longread sequencing platforms SMRT Sequencing offers long reads, high accuarcy, uniform coverage, singlemolecule resolution, and epigenetics to drive discovery in life science.
SmartSeq and SmartSeq2 Switch Mechanism at the 5' End of RNA Templates SmartSeq was developed as a singlecell sequencing protocol with improved read coverage across transcripts Complete coverage across the genome allows the detection of alternative transcript isoforms and SNPs There are 2 versions of SmartSeq SmartSeq and Smartseq2. The SMARTSeq HT Kit is designed to generate highquality, fulllength cDNA directly from 1–100 cells or 10 pg–1 ng of total RNA in a highthroughput manner The kit is evolved from the SMARTSeq v4 Ultra Low Input RNA Kit for Sequencing (SMARTSeq v4) and features a simplified protocol, which combines reverse transcription and PCR. #Introduction to the Smartseq2 Pipeline The Smartseq2 Single Sample pipeline is designed by the Data Coordination Platform (opens new window) of the Human Cell Atlas (opens new window) to process singlecell RNAseq (scRNAseq) data generated by Smartseq2 assays (opens new window)The pipeline's workflow is written in WDL, is freely available on GitHub (opens new window), and can be run by.
Also known as the ventral division of medial prefrontal cortex, 4). About Seurat Seurat is an R package designed for QC, analysis, and exploration of singlecell RNAseq data Seurat aims to enable users to identify and interpret sources of heterogeneity from singlecell transcriptomic measurements, and to integrate diverse types of singlecell data. Echo SystemEnhanced SMARTSeq v4 for RNA Sequencing Introduction As the cost of sequencing has continued to decline by orders of magnitude in the past ten years, scientists are enabled to ask deeper and more complex questions about transcriptomics Consequently, the variety, quantity, and demands of RNA sequencing experiments have all increased.
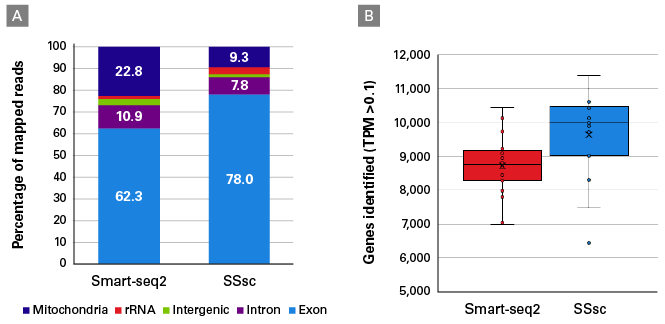
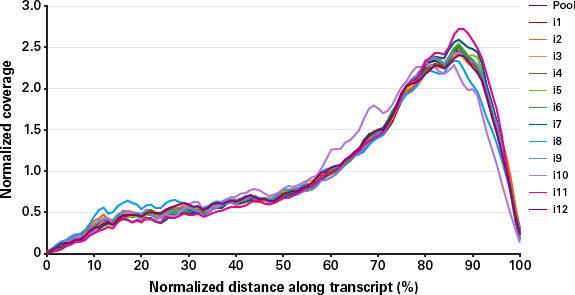
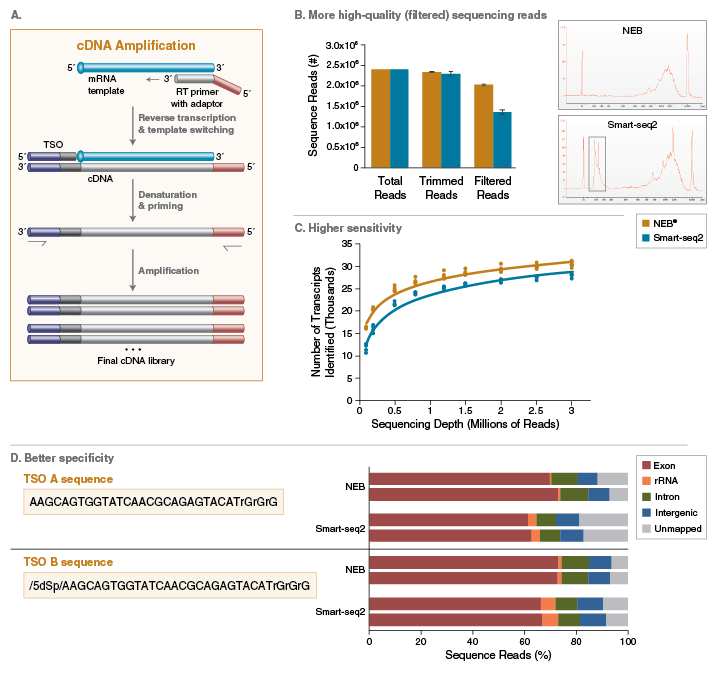
Echo SystemEnhanced SMARTSeq v4 for RNA Sequencing Introduction As the cost of sequencing has continued to decline by orders of magnitude in the past ten years, scientists are enabled to ask deeper and more complex questions about transcriptomics Consequently, the variety, quantity, and demands of RNA sequencing experiments have all increased. The NEBNext Single Cell/Low Input RNA Library Prep Kit, or the SMARTSeq v4 Ultra ® Low Input RNA Kit for Sequencing (Clontech ® # 6341) plus the Nextera XT DNA Library Prep Kit (Illumina ® #FC) were used Libraries were sequenced on an Illumina NextSeq ® 500 using pairedend mode (2x76 bp) Gene body coverage shown is an average. Smartseq3 This repository contains the scripts and pipelines used to process and analyse Smartseq3 libraries, as described in HagemannJensen et al https//doiorg//s We here provide the code to perform the following steps, that are expanded upon in the dedicated subfolders.
SmartSeq was developed as a singlecell sequencing protocol with improved read coverage across transcripts (Ramskold et al, 12) Complete coverage across the genome allows the detection of alternative transcript isoforms and SNPs There are 2 versions of SmartSeq SmartSeq and Smartseq2. SmartSeq is a Trademark by Takara Bio Usa, Inc, the address on file for this trademark is 1290 Terra Bella Avenue, Mountain View, CA. Note these data were included in the subsequent “Human Multiple Cortical Areas SMARTseq” data set, which can be found here This data set includes singlenucleus transcriptomes from 15,928 total nuclei derived from both frozen and neurosurgical human brain specimens, to survey cell type diversity in the human middle temporal gyrus ()Nuclei from 8 human tissue donors ranging in age.
SMARTSeq v4 Ultra Low Input RNA Kit for Sequencing (Cat Nos 6348, 6348, 6340, 6341, 6342, 6343, and 6344) is designed to generate high quality, fulllength cDNA directly from 1–1,000 cells or 10 pg–10 ng of total RNA, in a convenient input volume of 1–10 µl. While Smartseq2 detected the most genes per cell and across cells, CELseq2, Dropseq, MARSseq, and SCRBseq quantified mRNA levels with less amplification noise due to the use of unique molecular identifiers (UMIs). The SMARTSeq HT Kit is designed to generate highquality, fulllength cDNA directly from 1–100 cells or 10 pg–1 ng of total RNA in a highthroughput manner The kit is evolved from the SMARTSeq v4 Ultra Low Input RNA Kit for Sequencing (SMARTSeq v4) and features a simplified protocol, which combines reverse transcription and PCR.
Human V1 and ACC SMARTseq Note these data were included in the subsequent “Human Multiple Cortical Areas SMARTseq” data set, which can be found here This data set includes singlenucleus transcriptomes from 16,281 total nuclei collected from human primary visual cortex ( V1C ) and anterior cingulate cortex ( ACC ;. SMARTSeq v2, a method developed by Simone Picelli et al at the Karolinska Institute, has become a ubiquitous method for singlecell and population RNA sequencing Here, we demonstrate miniaturization of this process on human brain total RNA utilizing the Echo 525 Liquid Handler We show that this enhanced process can effectively reduce. SmartSeq and SmartSeq2 Switch Mechanism at the 5' End of RNA Templates SmartSeq was developed as a singlecell sequencing protocol with improved read coverage across transcripts Complete coverage across the genome allows the detection of alternative transcript isoforms and SNPs There are 2.
Also known as the ventral division of medial prefrontal cortex, 4). Smartseq3 is a unique quantitative fulllength scRNAseq protocol Smartseq3 uses the established 96 & 384 well microplate format for its library preparations To start, single cells must be. Here, we describe Smartseqtotal, a method capable of assaying a broad spectrum of coding and noncoding RNA from a single cell Built upon the templateswitch mechanism, Smartseqtotal bears the key feature of its predecessor, Smartseq2, namely, the ability to capture fulllength transcripts with high yield and quality.
Single cell sequencing examines the sequence information from individual cells with optimized nextgeneration sequencing (NGS) technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment For example, in cancer, sequencing the DNA of individual cells can give information about. Allen Cell Types Database Your web browser does not meet one or more of the system requirements for this site. Overview of SmartSeq protocol SmartSeq cDNA illumina Nextera XT • Fragment cDNA and add sequencing adapters • Pairedend sequencing recommended (unstranded library) • ~3040 million reads per sample recommended.
I am currently trying to run both your scTransform and standard workflow to integrate some datasets that were generated with SmartSeq2 and 10X platforms For the SmartSeq2 datasets, what form should I upload the gene expression data as (.
Generation Of Count Matrix Introduction To Single Cell Rna Seq

Smartseq Ngs Bioinformatics Services
Http Genome Cshlp Org Content 29 11 1816 Full Pdf
Smart Seq のギャラリー
Http Www Takarabio Com Resourcedocument X
Streamlined Single Cell Mrna Seq Smart Seq Ht

Comparative Analysis Of Single Cell Rna Sequencing Methods Biorxiv

Low Input And Low Bias Ion Torrent Mrna Seq A Principle Of The Download Scientific Diagram
Www Takarabio Com Resourcedocument X
Analysis Of Single Cell Rna Seq Data 18 Bioinfosummer Workshop

The Promise Of Single Cell Rna Sequencing For Kidney Disease Investigation Abstract Europe Pmc
Www Bioconductor Org Help Course Materials 19 Csama Materials Lectures Csama19 Smartseq2 Pdf
Smart Seq Single Cell Kit Full Length Transcriptome Analysis With Ultimate Sensitivity For Single Cells
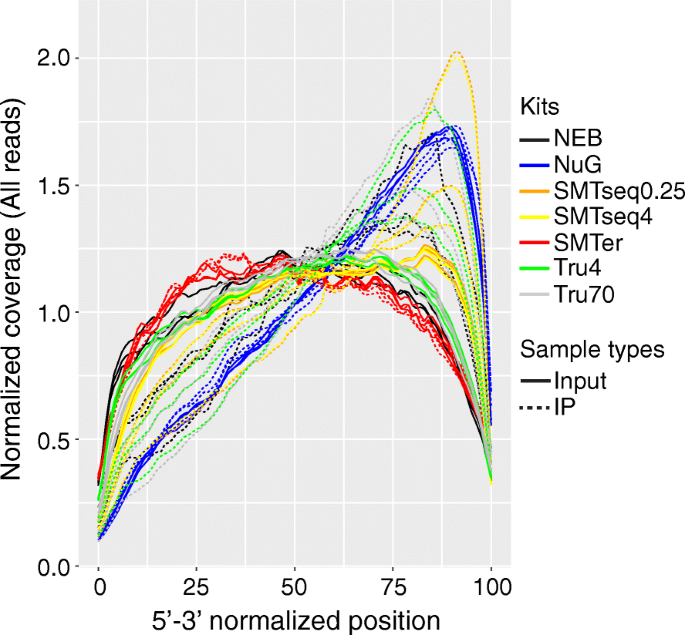
A Comparative Analysis Of Library Prep Approaches For Sequencing Low Input Translatome Samples Springerlink

Comparative Analysis Of Single Cell Rna Sequencing Methods Sciencedirect
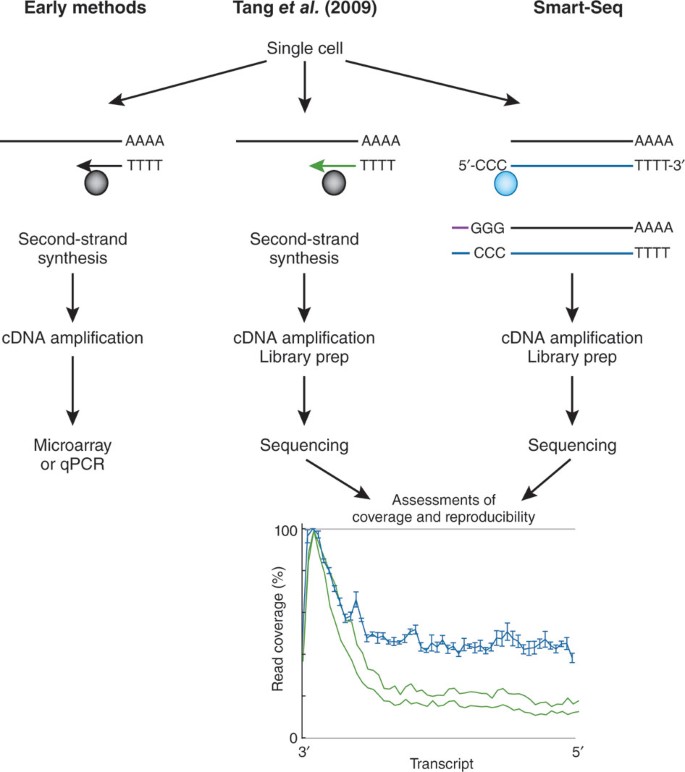
Transcriptome Sequencing Of Single Cells With Smart Seq Nature Biotechnology

Smart Seq Stranded Kit User Manual Pdf Free Download
Http Barc Wi Mit Edu Education Hot Topics Scrnaseq March19 Singlecellrnaseq Pdf

Yeast Single Cell Rna Seq Cell By Cell And Step By Step
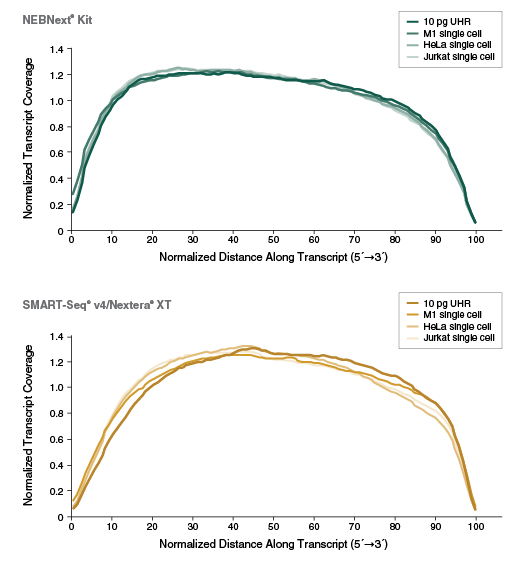
Overview Nebnext Single Cell Low Input Rna Library Prep Kit For Illumina Bioke
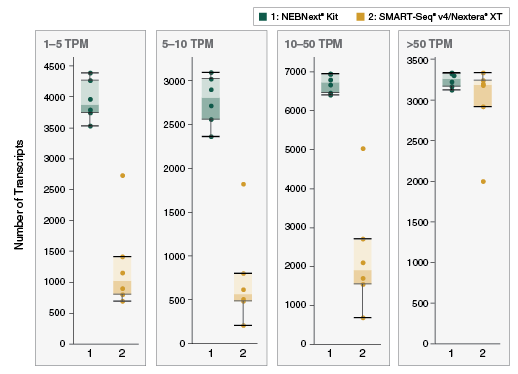
Overview Nebnext Single Cell Low Input Rna Library Prep Kit For Illumina Bioke
Q Tbn And9gcskgx7alacdunnntml64tiepz8nszabaaqacaz Ugzcj8vo7et4 Usqp Cau
2
Q Tbn And9gcqdogfgvb8ceank1 Lso0exsykd5gfzmn58g0fte9uk Hhptvbu Usqp Cau

Single Cell Profiling Of Total Rna Using Smart Seq Total Biorxiv
모바일 브릭
Smart Seq Single Cell Kit Full Length Transcriptome Analysis With Ultimate Sensitivity For Single Cells

Application Note Smart Seq Ht Protocol Namocell
Www Takarabio Com Resourcedocument X
Desktop Microfluidics For Low Input Rna Seq Sample Prep

Preparing Single Cell Library For Cheap Rna Seq Protocols That Work

Smart Seq Total Rna Seq Blog
Differential Expression From Single Cells Using The Smart Seq V4 3 De Kit

Smart Seq2
Www Takarabio Com Resourcedocument X

Smart 3seq A Fast Cheap Sensitive Rna Seq Protocol Part 1 Rna Seq Blog
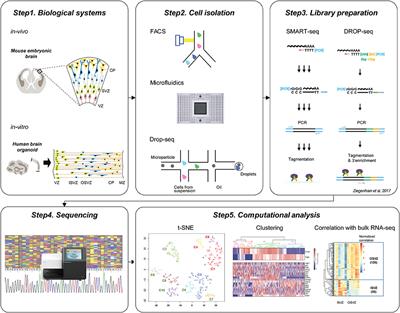
Frontiers Exploring The Complexity Of Cortical Development Using Single Cell Transcriptomics Neuroscience
Q Tbn And9gctoe5e6bkkiezmajyqyxkyp4qxafrh64tj2i49ppio Usqp Cau

Comparative Analysis Of Single Cell Rna Sequencing Methods Biorxiv

Identification Of Biomarkers For Amyotrophic Lateral Sclerosis By Comprehensive Analysis Of Exosomal Mrnas In Human Cerebrospinal Fluid Bmc Medical Genomics Full Text
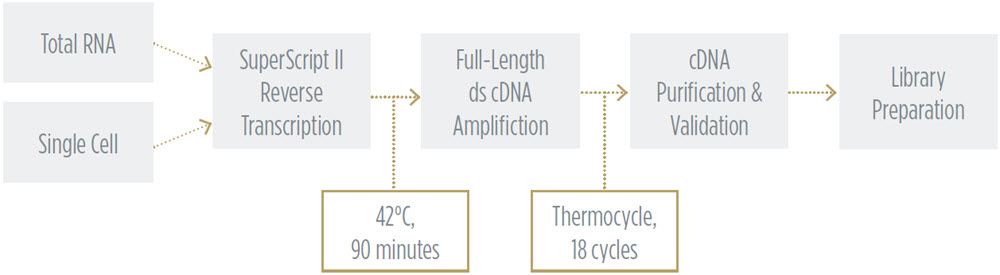
Echo System Enhanced Smart Seq V2 Rna Sequencing Beckman Coulter

3 Diagram Of Smart Seq Method After Cell Lysis Mrnas Are Download Scientific Diagram

High Resolution Genome Wide Expression Analysis Of Single Myofibers Using Smart Seq Sciencedirect
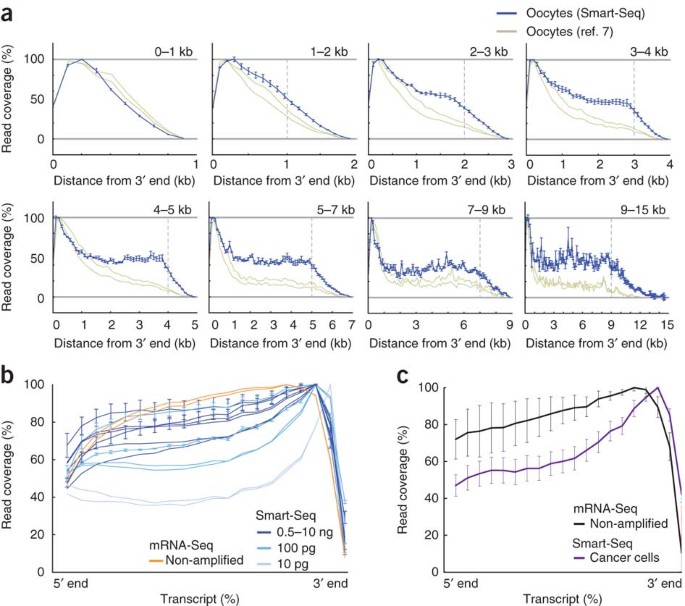
Full Length Mrna Seq From Single Cell Levels Of Rna And Individual Circulating Tumor Cells Nature Biotechnology
Www Takarabio Com Resourcedocument X1070

Single Cell Rna Sequencing Reagent Kit Smart Seq V4 Ultra Takara For Dna Sequencing Low Range

Pin On Rna Seq

Single Cell Sequencing For Precise Cancer Research Progress And Prospects Cancer Research

Sensitivity And Variability In Smart Seq From Few Or Single Cells A Download Scientific Diagram
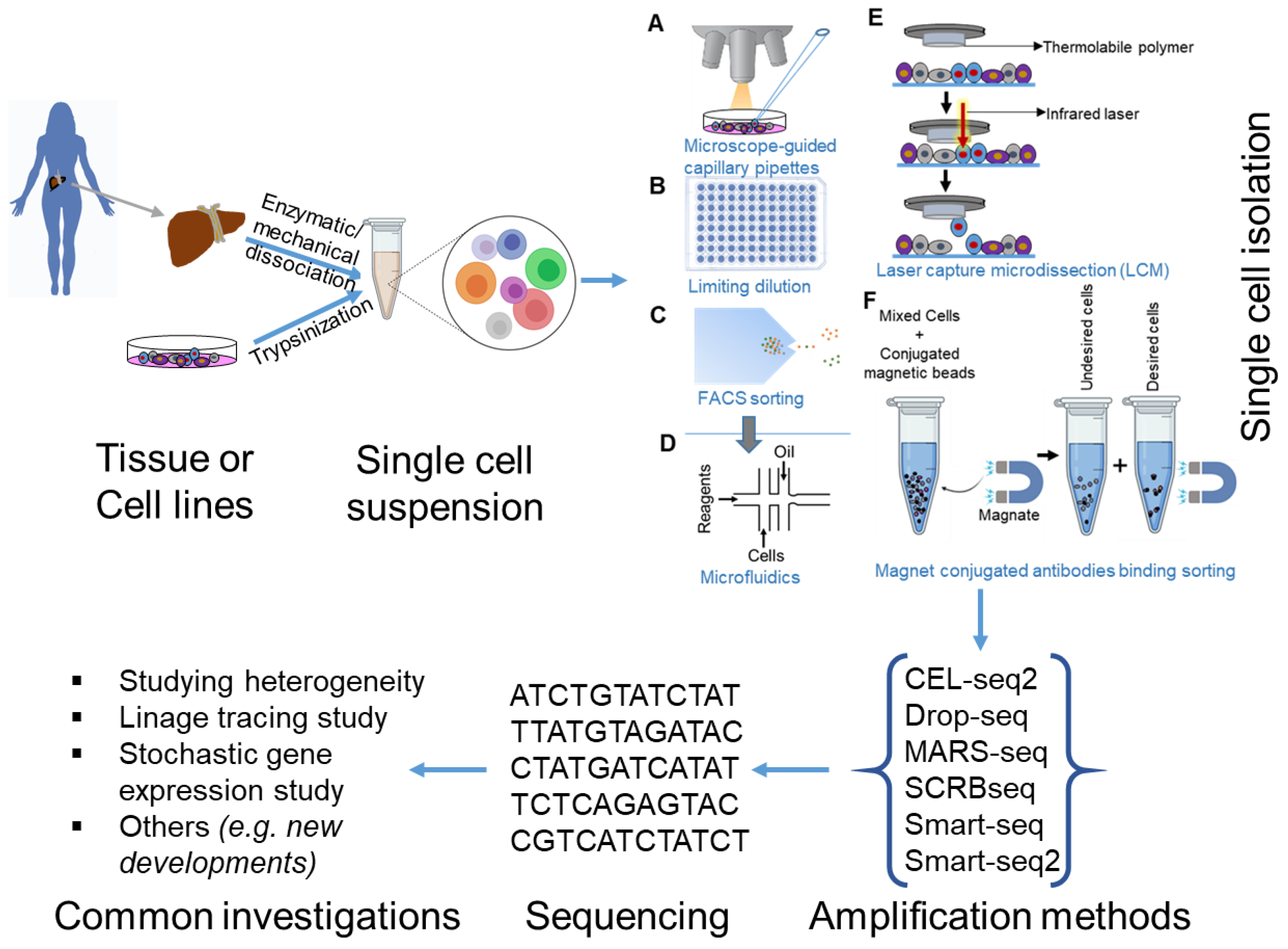
Cells Free Full Text Biological And Medical Importance Of Cellular Heterogeneity Deciphered By Single Cell Rna Sequencing Html
Www Fluidigm Com Binaries Content Documents Fluidigm Marketing Single Cell Genomics Polaris Deck Single Cell Genomics Polaris Deck Fluidigm 3afile

Comparative Analysis Of Single Cell Rna Sequencing Methods Biorxiv

Pdf Single Cell Profiling Of Total Rna Using Smart Seq Total

Buffer Solution Reagent Kit Smart Seq Ht Takara For Dna Sequencing For Single Cell Rna Sequencing For Nucleic Acids
Rna Seq Wikipedia
Apexbio Smart Seq2 Single Cell Kit

Transcriptome Sequencing Of Single Cells With Smart Seq Semantic Scholar

Rna Seq From Single Cells Archive Seqanswers
Www Bioconductor Org Help Course Materials 19 Csama Materials Lectures Csama19 Smartseq2 Pdf

Introduction To Single Cell Rna Seq
Template Switching Rt Enzyme Mix Bioke
Www Cytena Com Wp Content Uploads 07 Cytena F Sight I Dot Single Cell Omics App Note Pdf
Differential Expression From Single Cells Using The Smart Seq V4 3 De Kit
Full Length Single Cell Rna Sequencing With Smart Seq2 Springerlink

Flowchart For Smart Seq2 Library Preparation Outline Of The Protocol Download Scientific Diagram
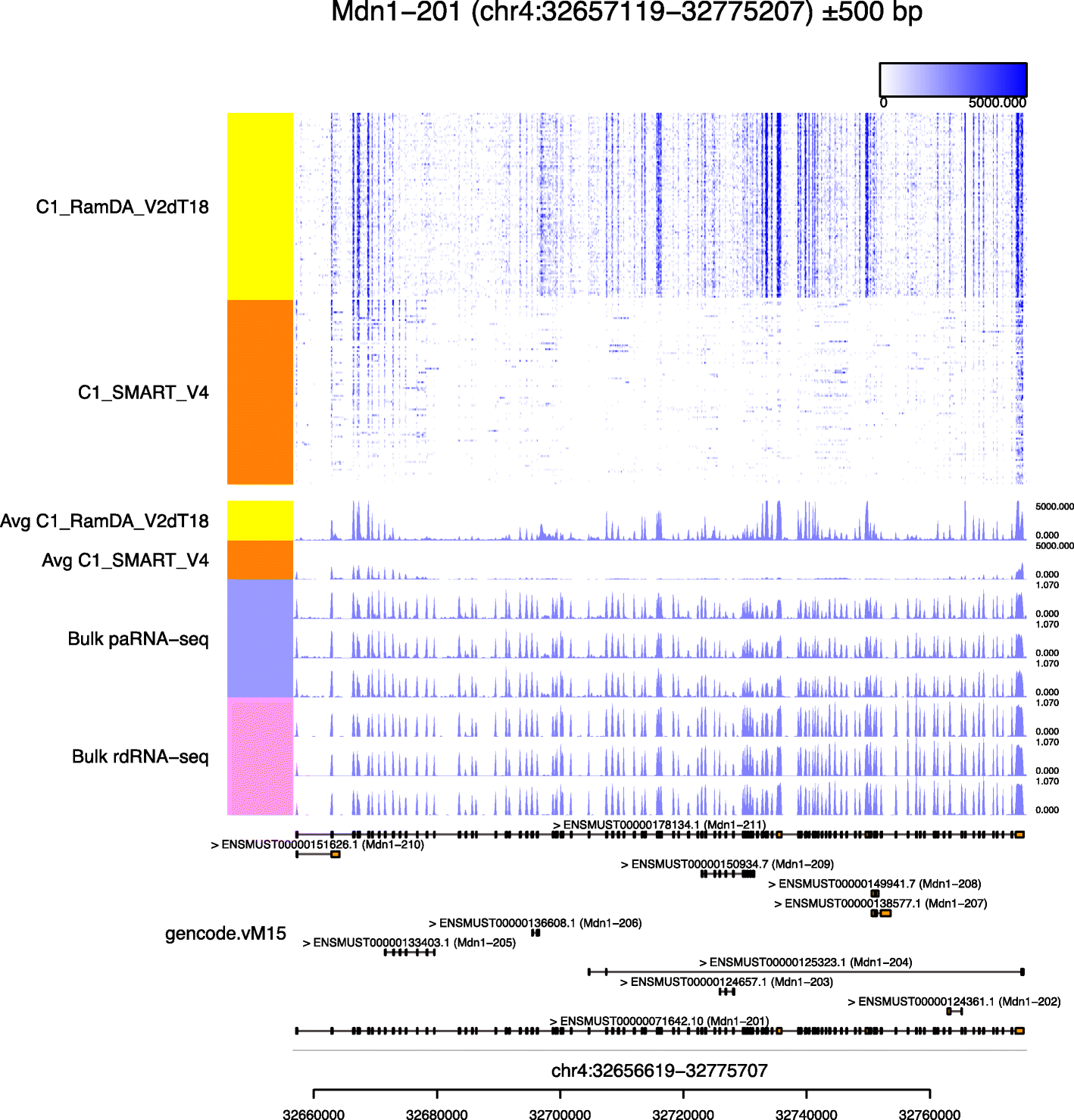
Millefy Visualizing Cell To Cell Heterogeneity In Read Coverage Of Single Cell Rna Sequencing Datasets Bmc Genomics Full Text

Smart Seq V4 Ultra Low Input Rna Kit From Takara Bio Selectscience

Comparative Analysis Of Single Cell Rna Sequencing Methods Sciencedirect
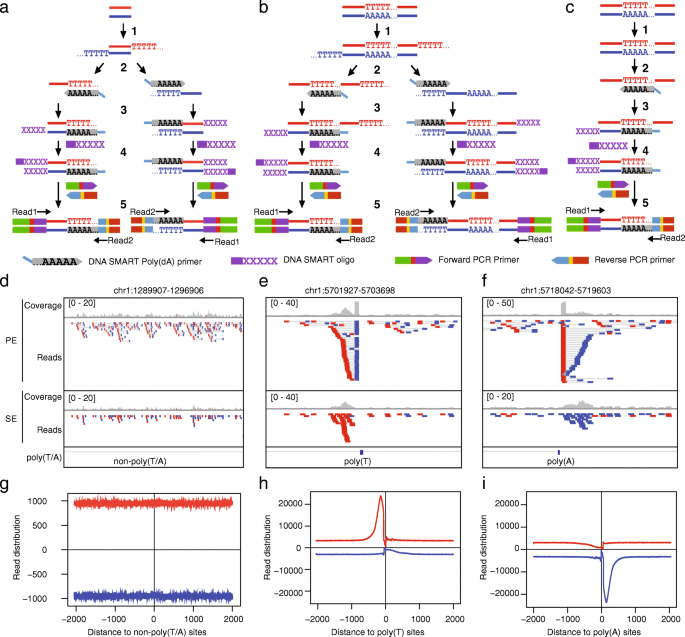
Smartcleaner Identify And Clean Off Target Signals In Smart Chip Seq Analysis Bmc Bioinformatics Full Text

Namocell S Smart Seq Ht Protocol Bioke

Smart Seq2 Rna Seq Blog
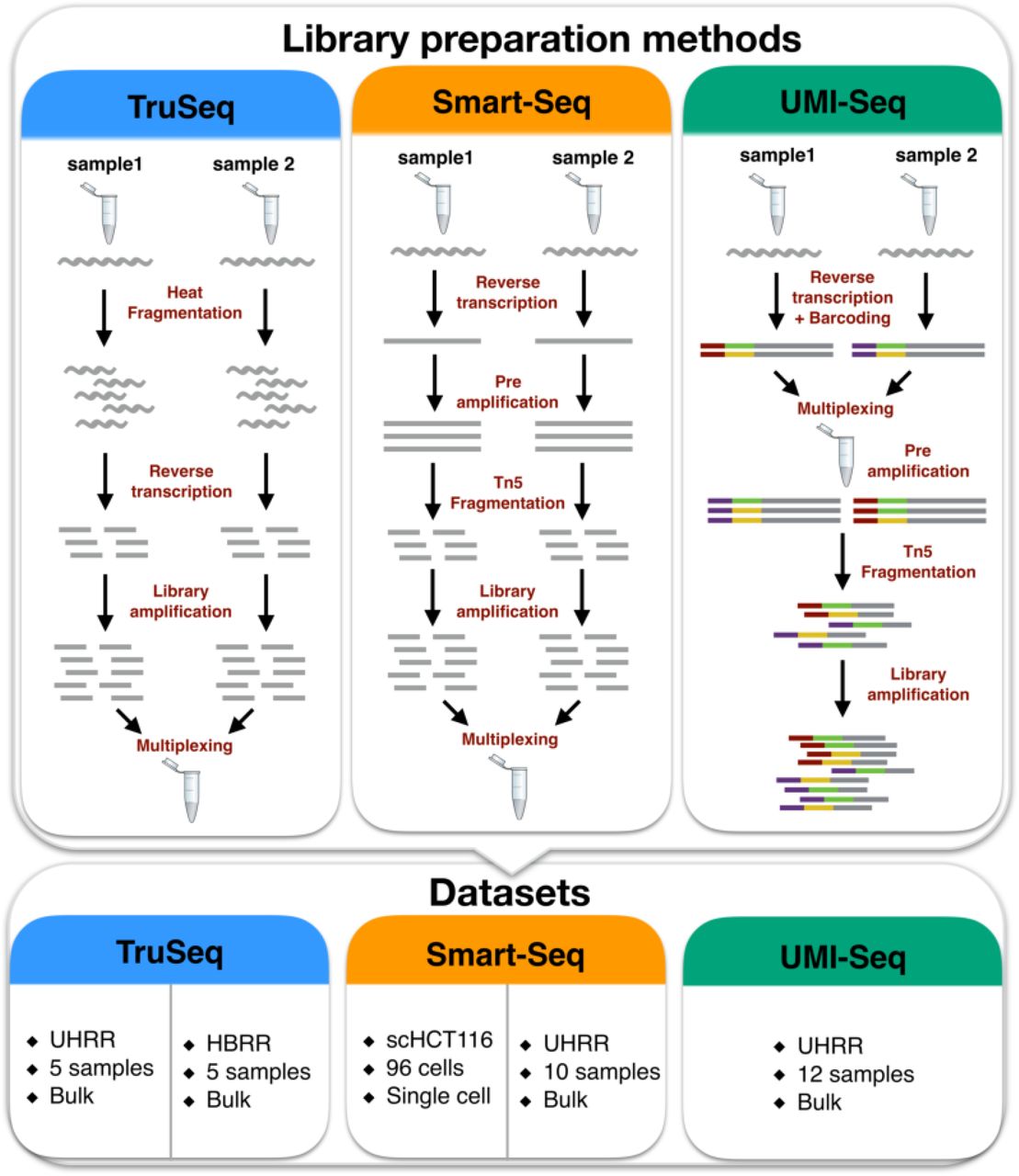
The Impact Of Amplification On Differential Expression Analyses By Rna Seq Biorxiv
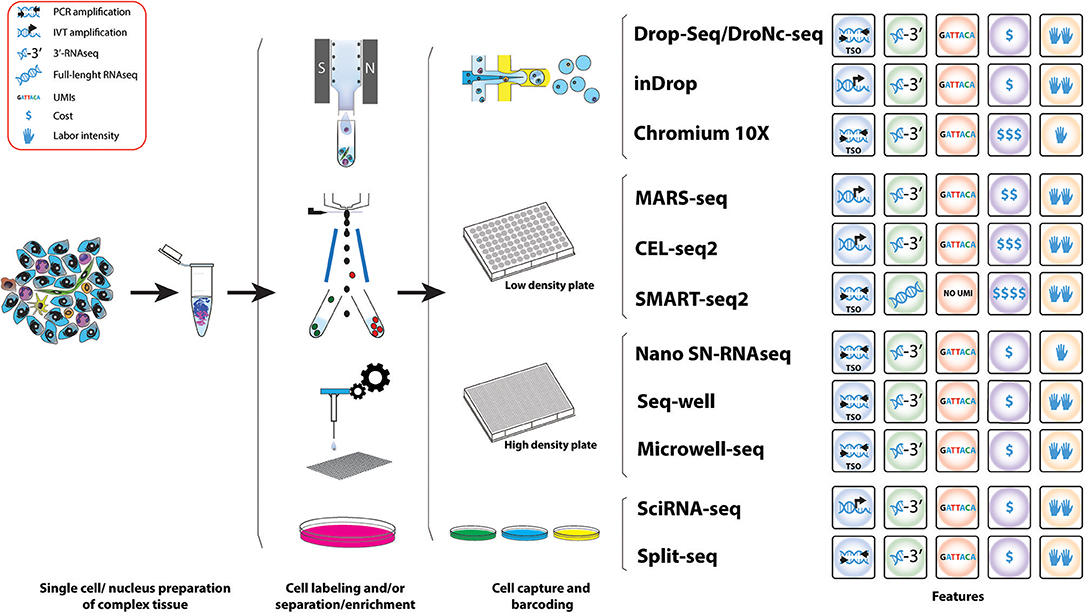
Frontiers Single Cell Transcriptomics In Cancer Immunobiology The Future Of Precision Oncology Immunology
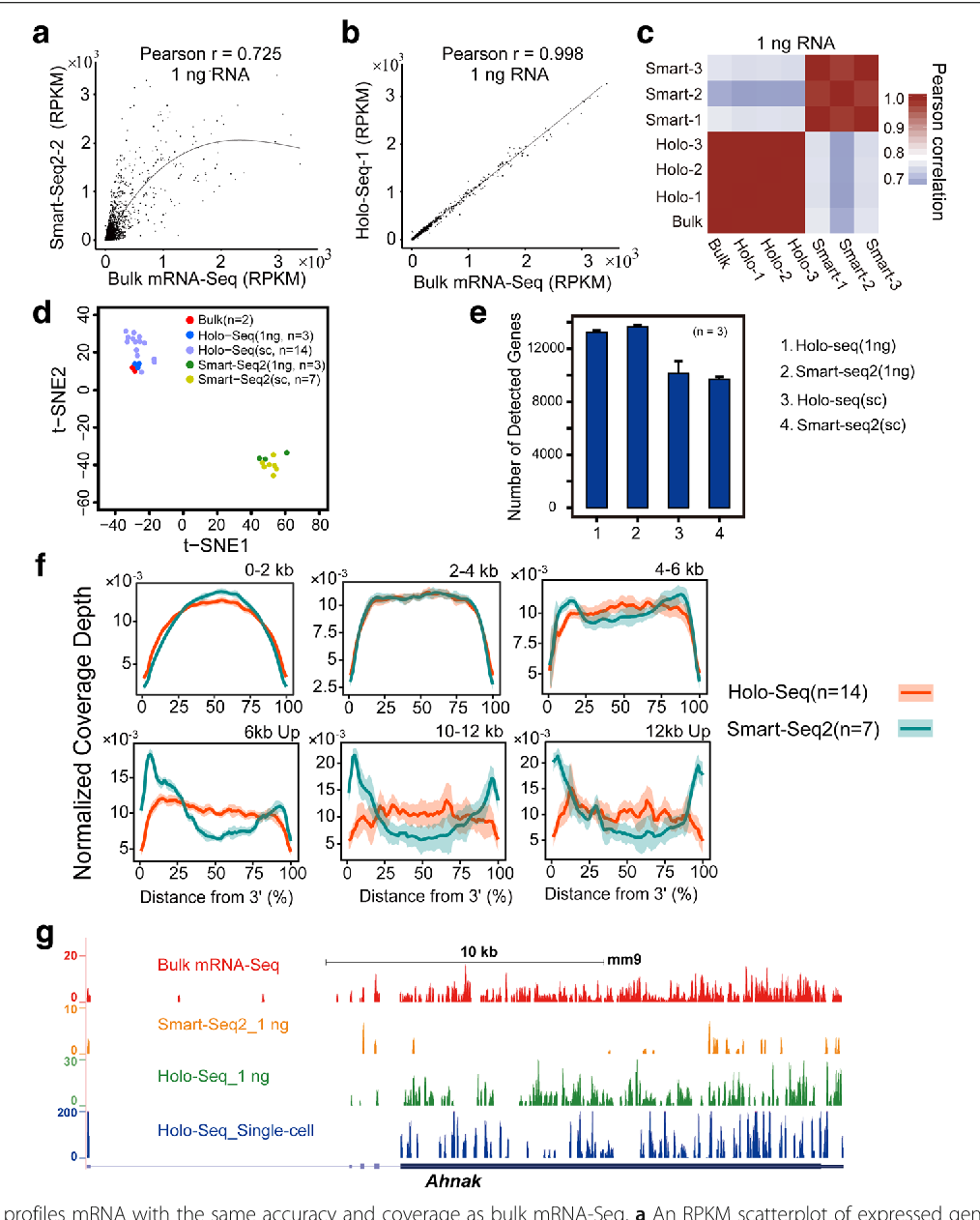
Figure 1 From Holo Seq Single Cell Sequencing Of Holo Transcriptome Semantic Scholar

Single Cell Smart Seq Data A Representative Images Of 1 Cell Download Scientific Diagram

Unravelling Intratumoral Heterogeneity Through High Sensitivity Single Cell Mutational Analysis And Parallel Rna Sequencing Sciencedirect

Pdf Full Length Rna Seq From Single Cells Using Smart Seq2

Smart Seq2 Single Cell Rna Seq Modified Method
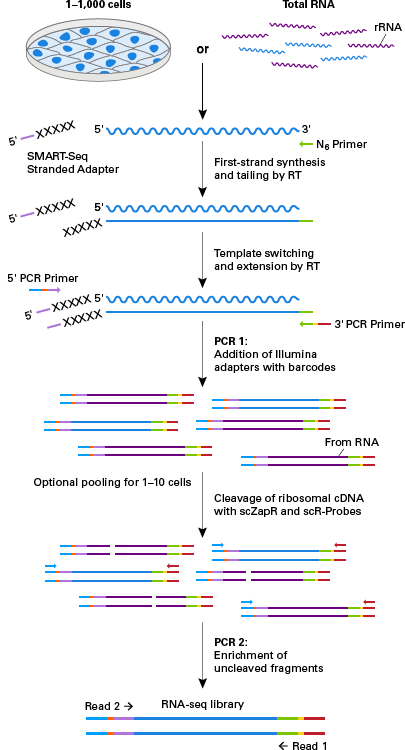
High Quality Stranded Rna Seq Libraries From Single Cells Using The Smart Seq Stranded Kit
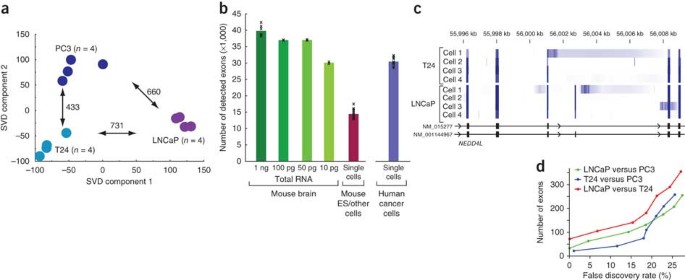
Full Length Mrna Seq From Single Cell Levels Of Rna And Individual Circulating Tumor Cells Nature Biotechnology
Www Biorxiv Org Content 10 1101 v1 Full Pdf

Identification Of Cell Types Using Scrna Seq Data From Smart Seq2 Download Scientific Diagram

Smart Seq Rna Seq Blog

Figure Coverage Sensitivity And Accuracy Of Matq Seq Compared With Download Scientific Diagram
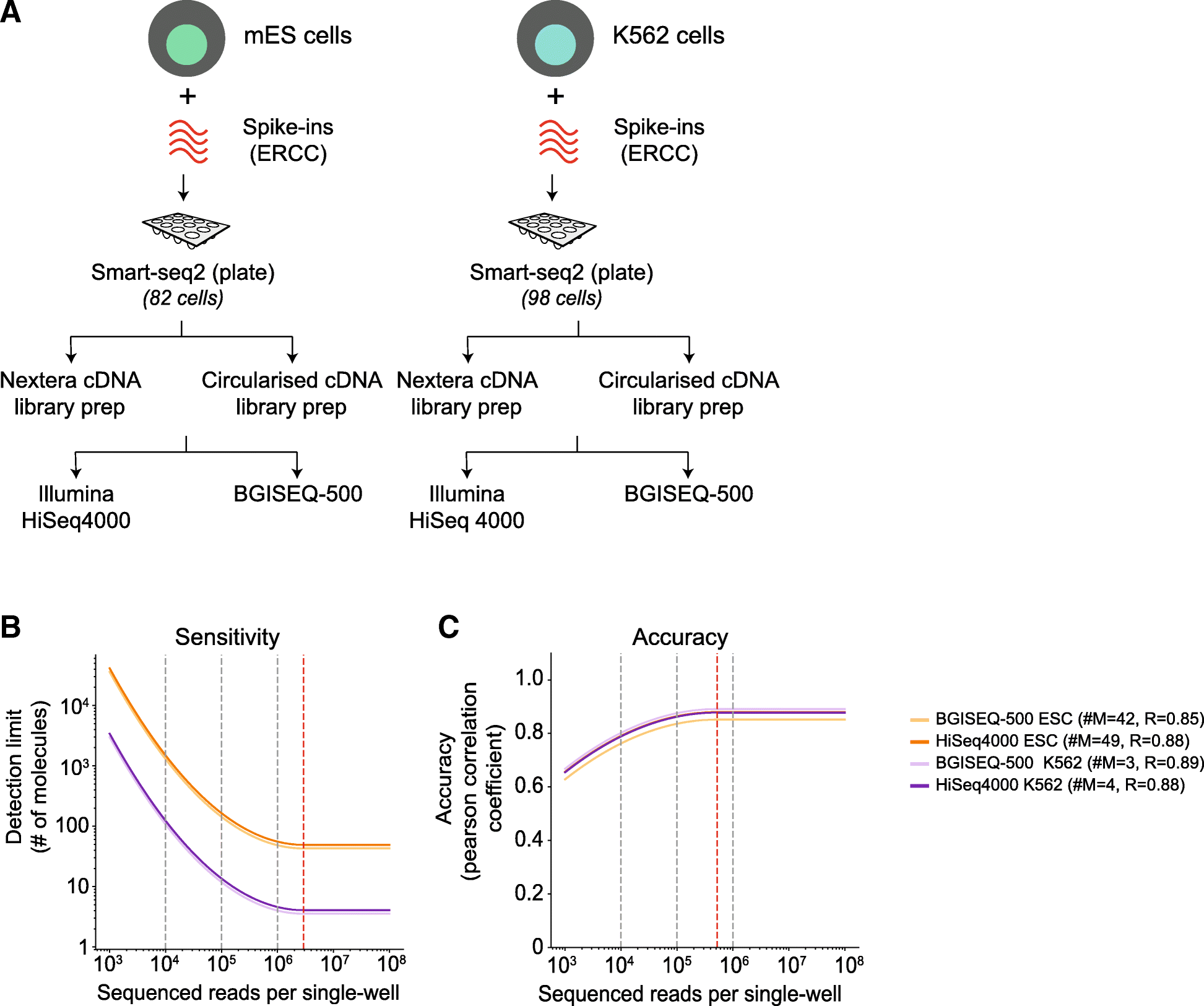
Comparative Analysis Of Sequencing Technologies For Single Cell Transcriptomics Genome Biology Full Text
Catalog Takara Bio Co Jp Pdfs Unprecedented Sensitivity With The Smart Seq Single Cell Kit Pdf

Comparative Analysis Of Single Cell Rna Sequencing Methods Sciencedirect

Smart Seq Total Rna Seq Blog

Comparison Of 10x And Smart Seq2 A The Protocols Differ In The Download Scientific Diagram

In Situ Transcriptome Characteristics Are Lost Following Culture Adaptation Of Adult Cardiac Stem Cells Biorxiv

Adapting The Smart Seq2 Protocol For Robust Single Worm

High Resolution Mapping Of Cell Types With Improved Single Cell Sequencing Rna Seq Blog

Flowchart For Smart Seq2 Library Preparation Outline Of The Protocol Download Scientific Diagram
Smart Seq Single Cell Kit Full Length Transcriptome Analysis With Ultimate Sensitivity For Single Cells

Methods Smart Seq2 Escg Eukaryotic Single Cell Genomics

Smart Seq Plus Kits From Takarabiousa Dss Takara Bio India Pvt Ltd Facebook
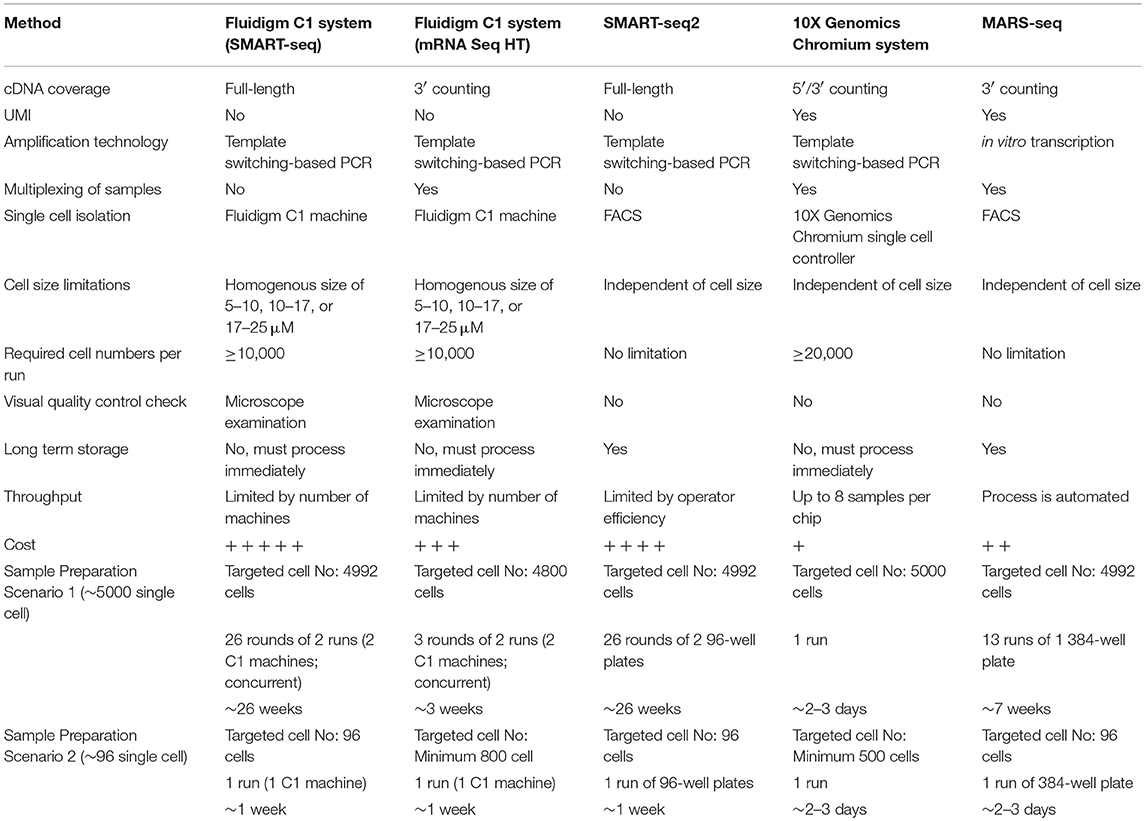
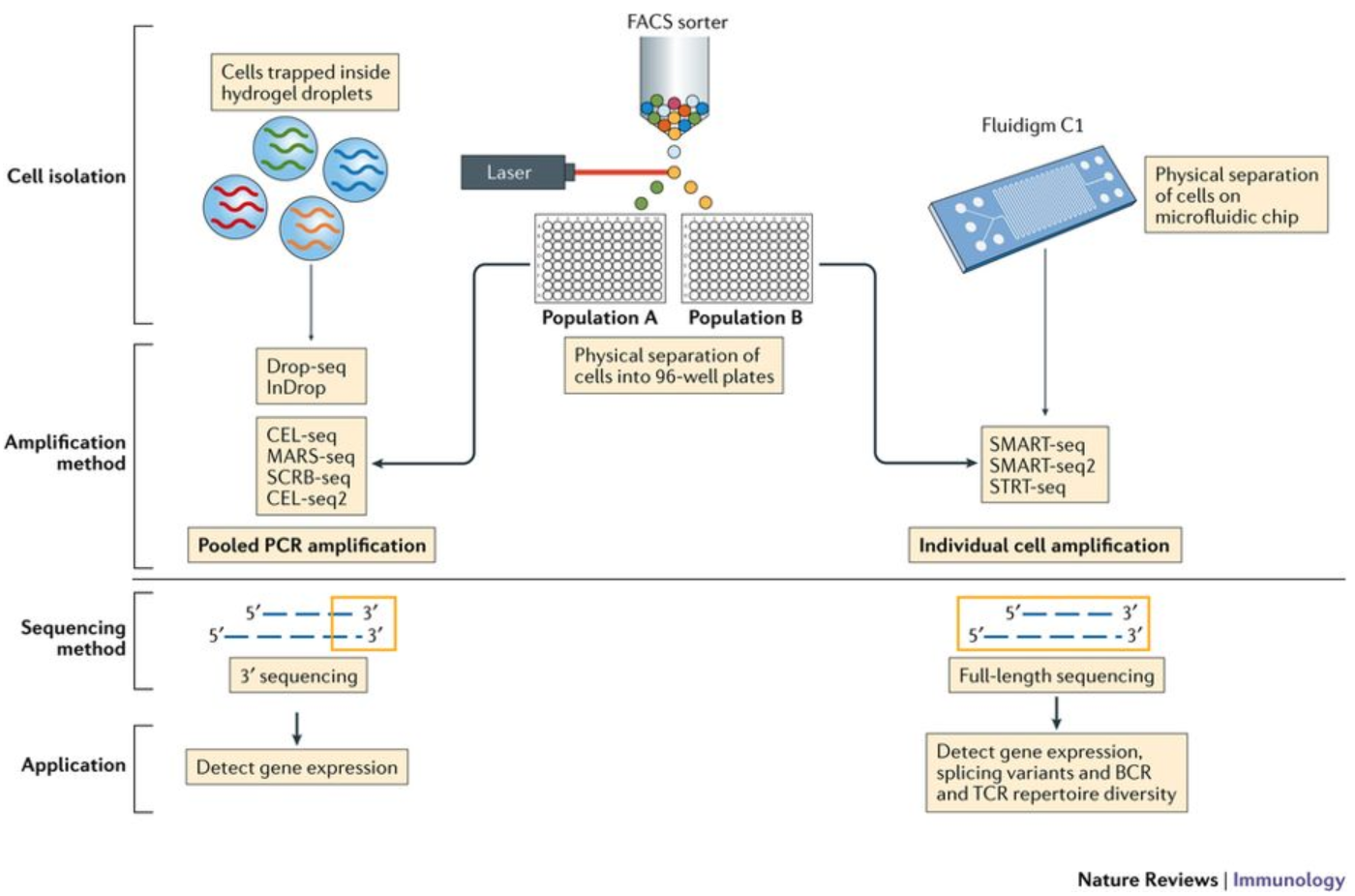
Frontiers A Single Cell Sequencing Guide For Immunologists Immunology
Q Tbn And9gcqj Finanaovbxxu58qm2wcnkqes9pkyccg7rjuvpru7jg1sz Usqp Cau

Comparative Analysis Of Single Cell Rna Sequencing Methods Sciencedirect

Single Cell Rna Seq Tutorial
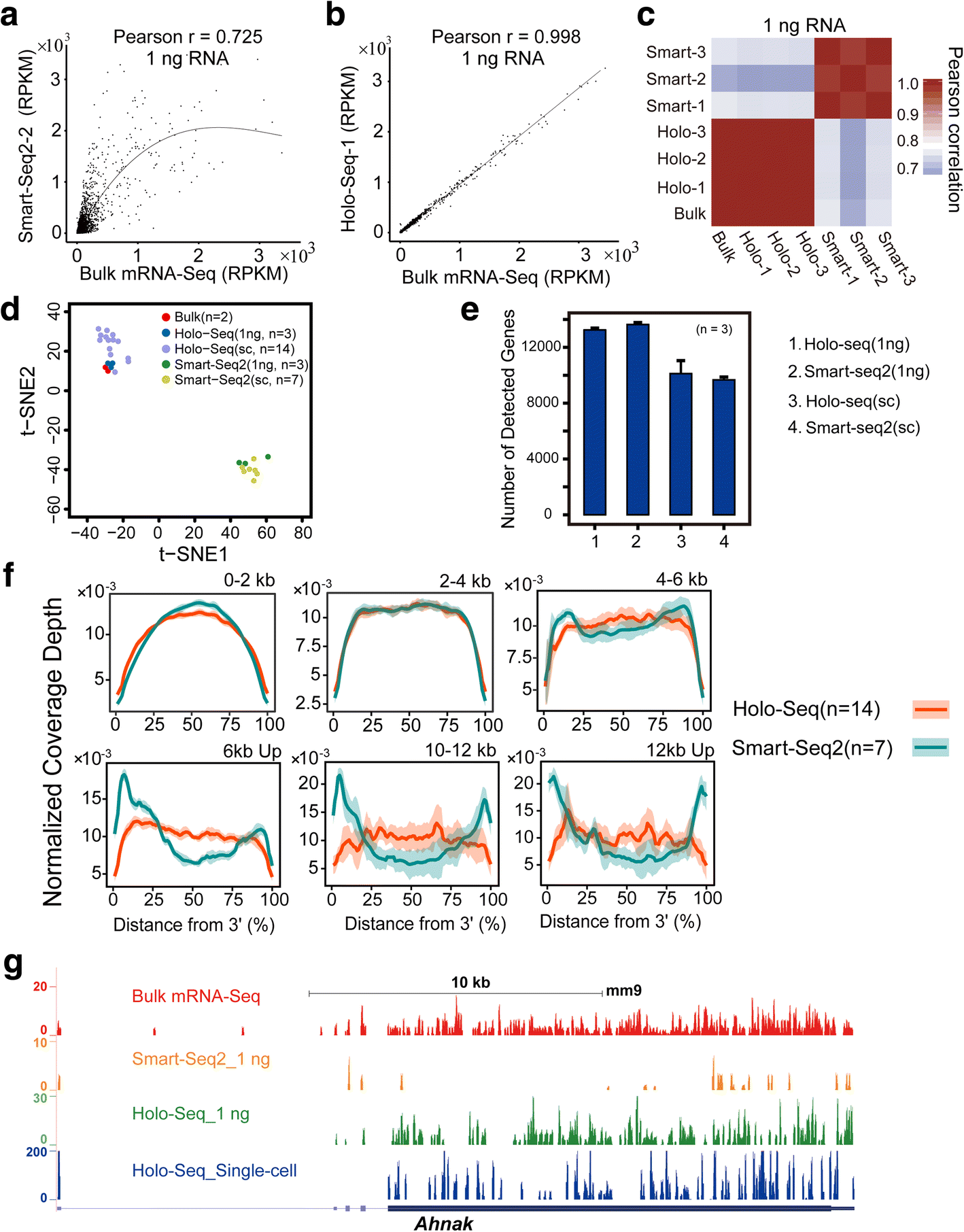
Holo Seq Single Cell Sequencing Of Holo Transcriptome Genome Biology Full Text

Help With Smart Seq Single Cell Rna Analysis Bioinformatics

Tradeoff Between More Cells And Higher Read Depth For Single Cell Rna Seq Spatial Ordering Analysis Of The Liver Lobule Biorxiv

Nature Protocols Rna Seq Blog

Unravelling Intratumoral Heterogeneity Through High Sensitivity Single Cell Mutational Analysis And Parallel Rna Sequencing Sciencedirect




